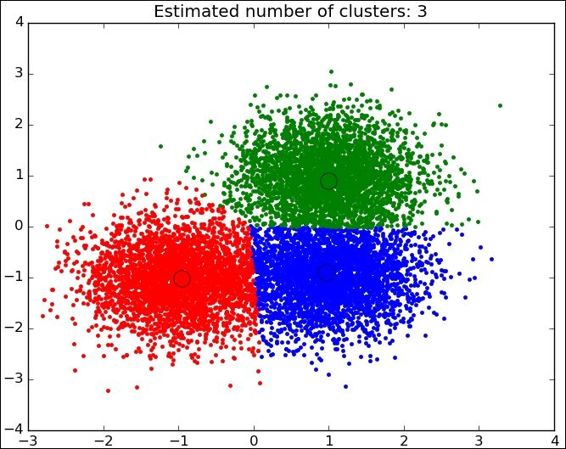
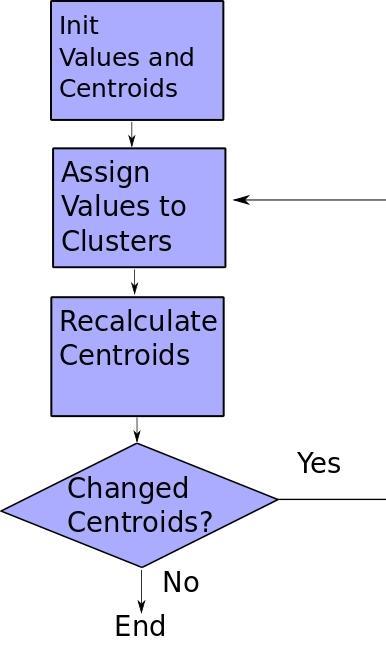
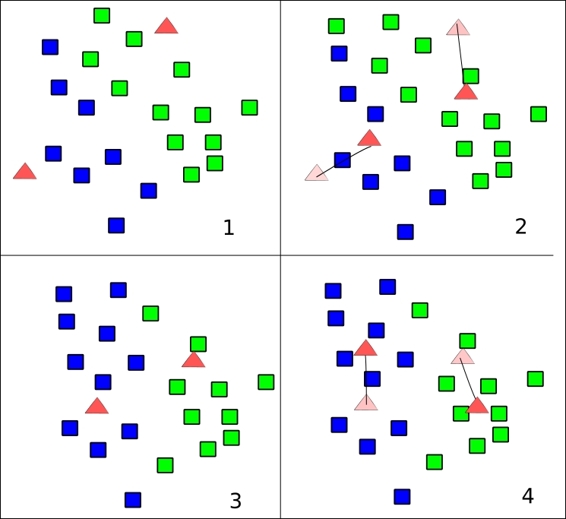
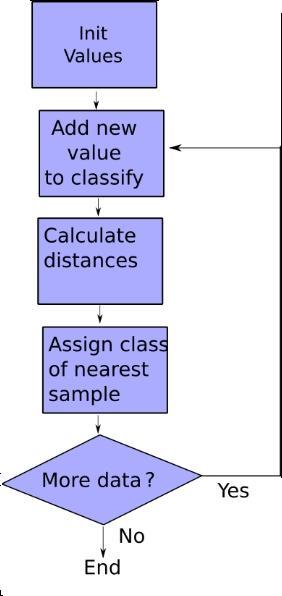
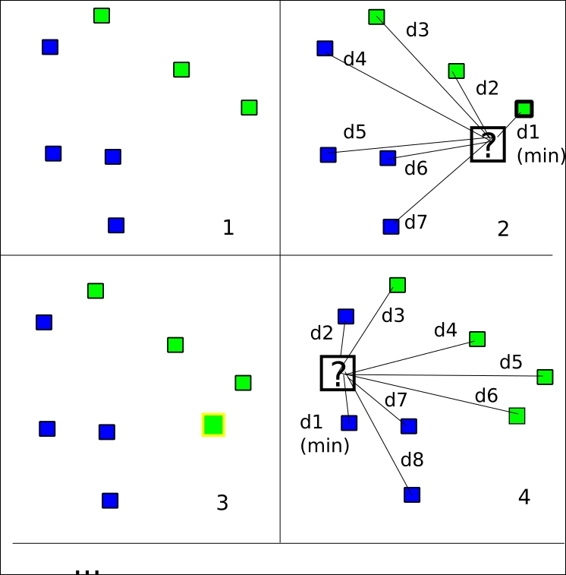
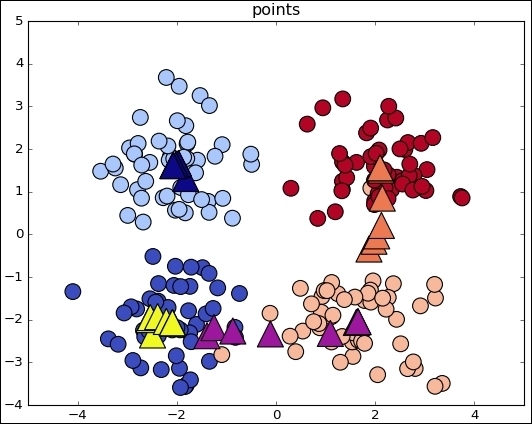
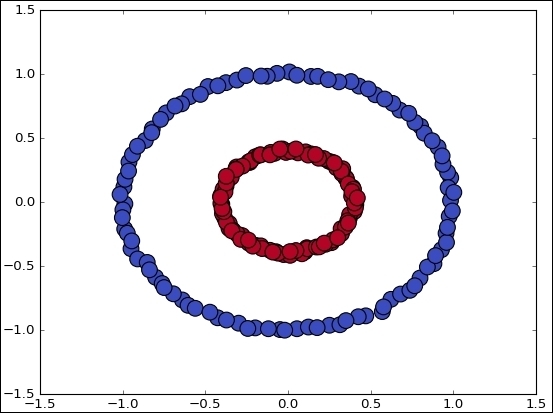
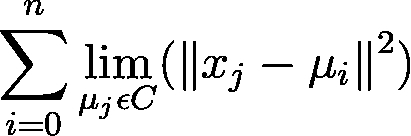Following is the complete source code:
import tensorflow as tf
import numpy as np
import time
import matplotlib
import matplotlib.pyplot as plt
from sklearn.datasets.samples_generator import make_blobs
from sklearn.datasets.samples_generator import make_circles
DATA_TYPE = 'blobs'
# Number of clusters, if we choose circles, only 2 will be enough
if (DATA_TYPE == 'circle'):
K=2
else:
K=4
# Maximum number of iterations, if the conditions are not met
MAX_ITERS = 1000
start = time.time()
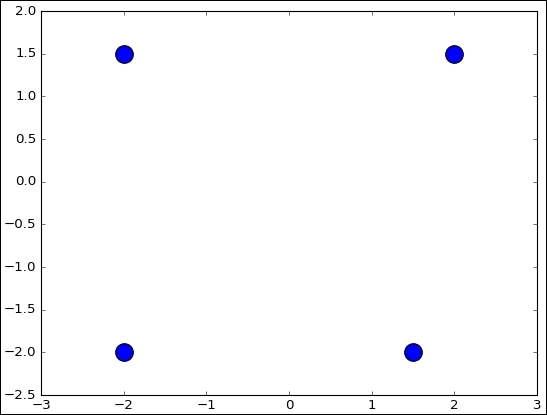
centers = [(-2, -2), (-2, 1.5), (1.5, -2), (2, 1.5)]
if (DATA_TYPE == 'circle'):
data, features = make_circles(n_samples=200, shuffle=True, noise= 0.01, factor=0.4)
else:
data, features = make_blobs (n_samples=200, centers=centers, n_features = 2, cluster_std=0.8, shuffle=False, random_state=42)
fig, ax = plt.subplots()
ax.scatter(np.asarray(centers).transpose()[0], np.asarray(centers).transpose()[1], marker = 'o', s = 250)
plt.show()
fig, ax = plt.subplots()
if (DATA_TYPE == 'blobs'):
ax.scatter(np.asarray(centers).transpose()[0], np.asarray(centers).transpose()[1], marker = 'o', s = 250)
ax.scatter(data.transpose()[0], data.transpose()[1], marker = 'o', s = 100, c = features, cmap=plt.cm.coolwarm )
plt.plot()
points=tf.Variable(data)
cluster_assignments = tf.Variable(tf.zeros([N], dtype=tf.int64))
centroids = tf.Variable(tf.slice(points.initialized_value(), [0,0], [K,2]))
sess = tf.Session()
sess.run(tf.initialize_all_variables())
rep_centroids = tf.reshape(tf.tile(centroids, [N, 1]), [N, K, 2])
rep_points = tf.reshape(tf.tile(points, [1, K]), [N, K, 2])
sum_squares = tf.reduce_sum(tf.square(rep_points - rep_centroids),
reduction_indices=2)
best_centroids = tf.argmin(sum_squares, 1)
did_assignments_change = tf.reduce_any(tf.not_equal(best_centroids, cluster_assignments))
def bucket_mean(data, bucket_ids, num_buckets):
total = tf.unsorted_segment_sum(data, bucket_ids, num_buckets)
count = tf.unsorted_segment_sum(tf.ones_like(data), bucket_ids, num_buckets)
return total / count
means = bucket_mean(points, best_centroids, K)
with tf.control_dependencies([did_assignments_change]):
do_updates = tf.group(
centroids.assign(means),
cluster_assignments.assign(best_centroids))
changed = True
iters = 0
fig, ax = plt.subplots()
if (DATA_TYPE == 'blobs'):
colourindexes=[2,1,4,3]
else:
colourindexes=[2,1]
while changed and iters < MAX_ITERS:
fig, ax = plt.subplots()
iters += 1
[changed, _] = sess.run([did_assignments_change, do_updates])
[centers, assignments] = sess.run([centroids, cluster_assignments])
ax.scatter(sess.run(points).transpose()[0], sess.run(points).transpose()[1], marker = 'o', s = 200, c = assignments, cmap=plt.cm.coolwarm )
ax.scatter(centers[:,0],centers[:,1], marker = '^', s = 550, c = colourindexes, cmap=plt.cm.plasma)
ax.set_title('Iteration ' + str(iters))
plt.savefig("kmeans" + str(iters) +".png")
ax.scatter(sess.run(points).transpose()[0], sess.run(points).transpose()[1], marker = 'o', s = 200, c = assignments, cmap=plt.cm.coolwarm )
plt.show()
end = time.time()
print ("Found in %.2f seconds" % (end-start)), iters, "iterations"
print "Centroids:"
print centers
print "Cluster assignments:", assignments
This is the simplest case for observing the algorithm mechanics. When the data comes from the real world, the classes are normally not so clearly separated and it is more difficult to label the data samples.